The authenticity of the strain of Bacillus coagulans in LactoSpore® has been established through genotyping and phenotypic characterization.
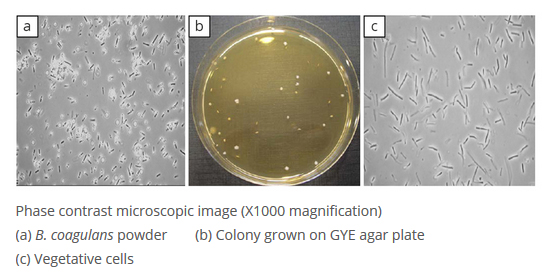
LactoSpore® contains a mixture of ellipsoidal terminal spores and vegetative cells (Fig. a). Colonies were grown on Glucose Yeast Extract (GYE) agar media, yielding uniform, 1–3 mm in diameter, white to cream, smooth colonies (Fig. b) that contain vegetative rod shaped cells (Fig. c). The results showed that all colonies isolated from five different production batch samples had an identical phenotype, consistent with the phenotype of B. coagulans.
Techniques such as 16s rRNA sequencing, GTG 5″, BOX-PCR fingerprinting and Multilocus sequence typing were carried out to evaluate the identity and consistency of LactoSpore®.
Two variations of repetitive intergenic DNA sequences (rep-PCR) genomic fingerprinting were performed using the BOX and GTG 5” primers. Analysis indicated that the sample contained the same strain through production batches. The isolated colonies from all five LactoSpore® production samples had identical BOX-PCR fingerprints, indicating that the strain present was B. coagulans, and that its identity was consistent.
Multilocus sequence typing performed on samples clearly indicated that the sequences from the LactoSpore® were all identical. The study indicated that there was no change in the household genes, which are far more sensitive to mutation than the 16S rDNA gene. This was yet another confirmation of strain purity and consistency over a period of time (Majeed et al., 2016).
